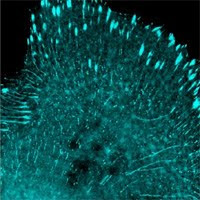
This week scientists from Allele Biotechnology and its partner non-profit research institute, the Scintillon Institute, present their latest fluorescent protein, mNeonGreen, in the journal Nature Methods (Nature Publishing Group). In the paper, entitled “A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum,” the scientists describe the development of the brightest monomeric fluorescent protein to date.
The
scientific efforts to develop this novel fluorescent protein were led
by Dr. Nathan Shaner, a leader in the field of fluorescent protein
engineering. Fluorescent proteins are highly valuable research tools
that allow the labeling and imaging of individual proteins within a
living cell, and tracking of their movements and localization in real
time through a microscope. However, since the discovery of the original
green fluorescent protein in 1993, imaging technology has advanced
rapidly beyond the capability of most fluorescent proteins. The newly
described fluorescent protein, mNeonGreen, allows researchers to take
full advantage of modern super-resolution optical microscopy techniques
that enable visualization of structures in living and fixed cells at
much smaller scales than are possible using traditional optical
microscopy. This improvement will lead to countless new insights into
human health and a greater understanding of protein interactions at very
small distance scales within living cells. According to Dr. Jiwu Wang,
the CEO of Allele Biotechnology, “Super-resolution imaging will become
the standard for publication in a short period of time, and mNeonGreen
allows researchers to meet this standard while still being compatible
with the equipment and methods they already use.”
Prominent
researchers within the fluorescent protein field are touting mNeonGreen
as a replacement for jellyfish-derived Aequorea GFP, one of the most
commonly used fluorescent proteins today. According to lead researcher
Dr. Nathan Shaner, “mNeonGreen can be directly substituted for other
green fluorescent proteins such as EGFP without the need for any
equipment changes,” making the upgrade an attractive prospect for many
researchers.
Allele
Biotechnology and Pharmaceuticals Inc. is a San Diego-based
biotechnology company specializing in the fields of RNAi, stem cells,
viral expression, camelid antibodies and fluorescent proteins. The
company has co-developed a number of fluorescent proteins
and other products for PALM or STORM super-resolution imaging 3D-SIM,
and STED imaging. With the arrival of mNeonGreen, Allele plans to
collaborate with leading imaging labs, microscope manufacturers, and
journals such as Nature Methods to further promote the
advantages and capabilities of the latest imaging methods. Additionally,
this announcement will coincide with the launch of a new super-resolution imaging web portal and plasmid depository via collaboration with the Scintillon Institute. The Scintillon Institute
is a non-profit research institute established in 2012 using seed
funding from Allele Biotech. The institute’s researchers are focused on
the development of biological tools to improve human health and quality
of life, including applications to cancer imaging, regenerative
medicine, and sustainable energy and food production.
For details about Allele’s new Superresolution FP distribution method, read our departmental and institutional usage page.