Reprogramming
of differentiated cells into induced pluripotent stem cells (iPSCs) is
commonly considered a stochastic process, i.e. with randomness, which
could offer a convenient excuse for the historical inefficiency and
inconstancy of making iPSCs. We have demonstrated time and again that
by using potent mRNA cocktails, the majority of the fibroblasts seeded
in a well can be converted into pluripotent stage in a nearly
synchronized manner (Warren et al. 2012, Warren and Wang 2013,
and this Allele Picture Blog series). mRNA molecules can function
robustly yet transiently while avoiding the need of entering the
nucleus, a bottle-neck for all DNA-based vehicles.
Other
researchers are used to the idea of clonal expansion partly because
isolating iPSCs from “clones” was a common step during reprogramming
using viruses or other low efficiency methods, even though those clones
were not necessarily from single precursor cells. This week, the Allele
iPSC team developed a new way of managing our mRNA reprogramming that
allowed us to achieve clonal iPSCs that appear to be a lot purer and
more likely true clones compared to previous reports, without
compromising any of the main benefits of our protocol, e.g. feeder-free,
xeno-free, footprint-free, very fast and highly efficient. This work
is currently supported by an NIDA/NIH grant to Dr. Jiwu Wang at Allele
Biotech.
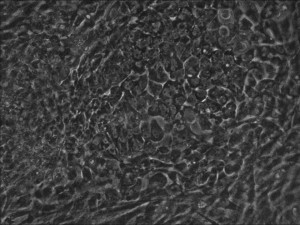
Traditional bulk-conversion by the Allele mRNA reprogramming protocol developed by Warren et al.
The picture shows large patches of cells becoming stem cells almost
overnight around the 9th day of adding mRNA-cocktail supplement to the
media.
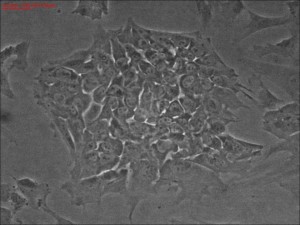
Clonal
iPSC formation using a modified mRNA reprogramming protocol. The
picture shows a typical clone of stem cells that originated from likely
single cells.
Warren, Ni, Wang, and Guo, Scientific Reports, 2012
Warren and Wang, Current Protocols, 2013, in press
Warren and Wang, Current Protocols, 2013, in press



No comments:
Post a Comment